Structural Characterization of the Molecules in the NDM Database.
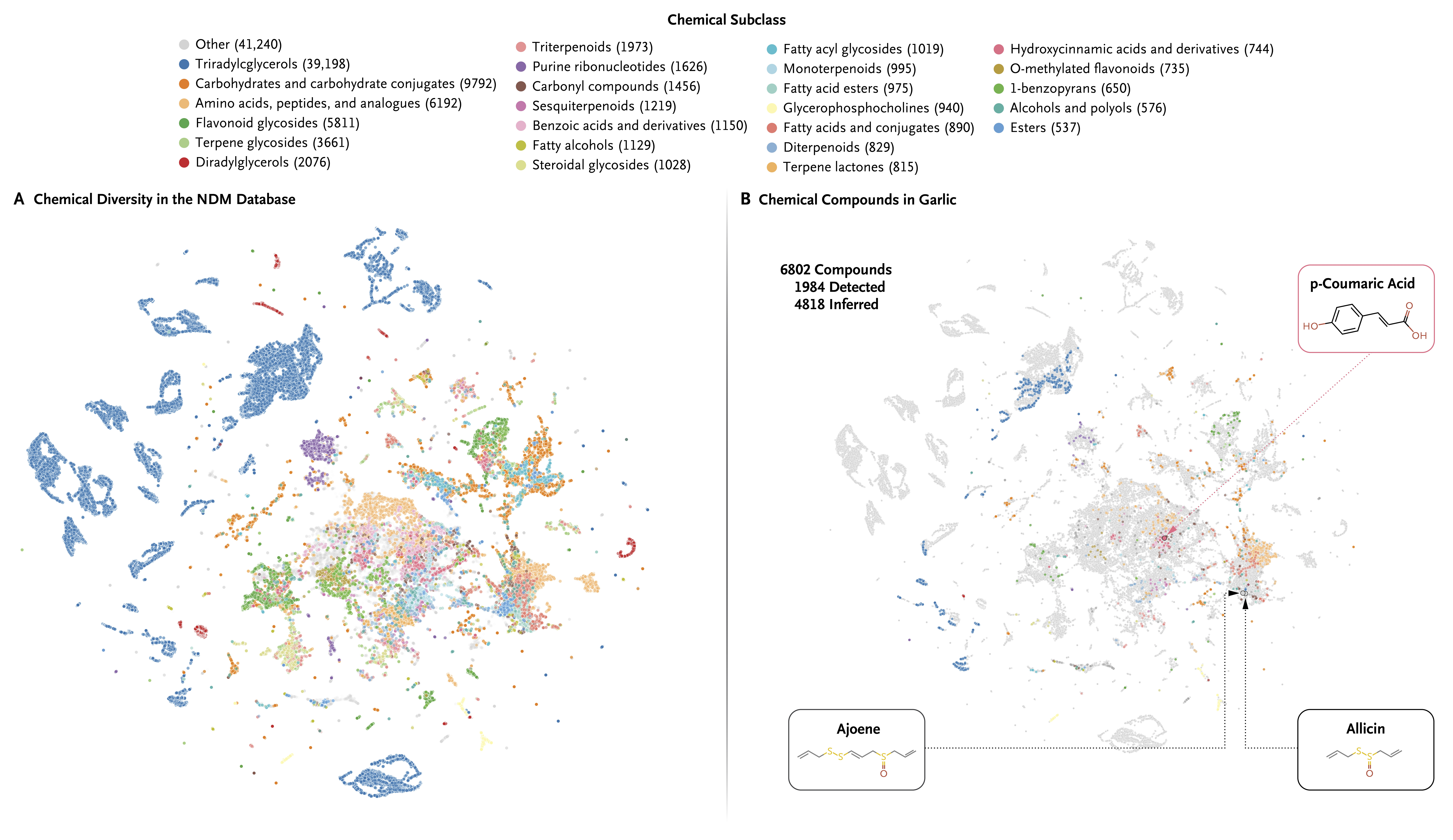
Panel A shows the chemical diversity in the Nutrition Dark Matter (NDM) database. With the use of contextual embedding vectors generated by the MoLFormer chemical language model and reduced to two dimensions with UMAP (Uniform Manifold Approximation and Projection), the space of food molecules in the NDM database can be visualized. Molecules are color-coded on the basis of structural subclasses (ClassyFire chemical taxonomy), each containing at least 500 compounds.
Panel B shows the chemical compounds in garlic. The same map highlights the 6802 molecules documented in garlic, categorized by structural subclasses. Three key chemical compounds — allicin and ajoene (organosulfur compounds) and p-coumaric acid (a polyphenol) — are emphasized for their relevance to human health, yet they are often overlooked in food composition databases.
Resources
Djoumbou Feunang Y, Eisner R, Knox C, et al. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J Cheminform 2016;8:61. [Medline]
McInnes L, Healy J, Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. Version 3. September 18, 2020 (https://arxiv.org/abs/1802.03426). preprint.
Comparison of Molecules in the NDM Database and DrugBank.
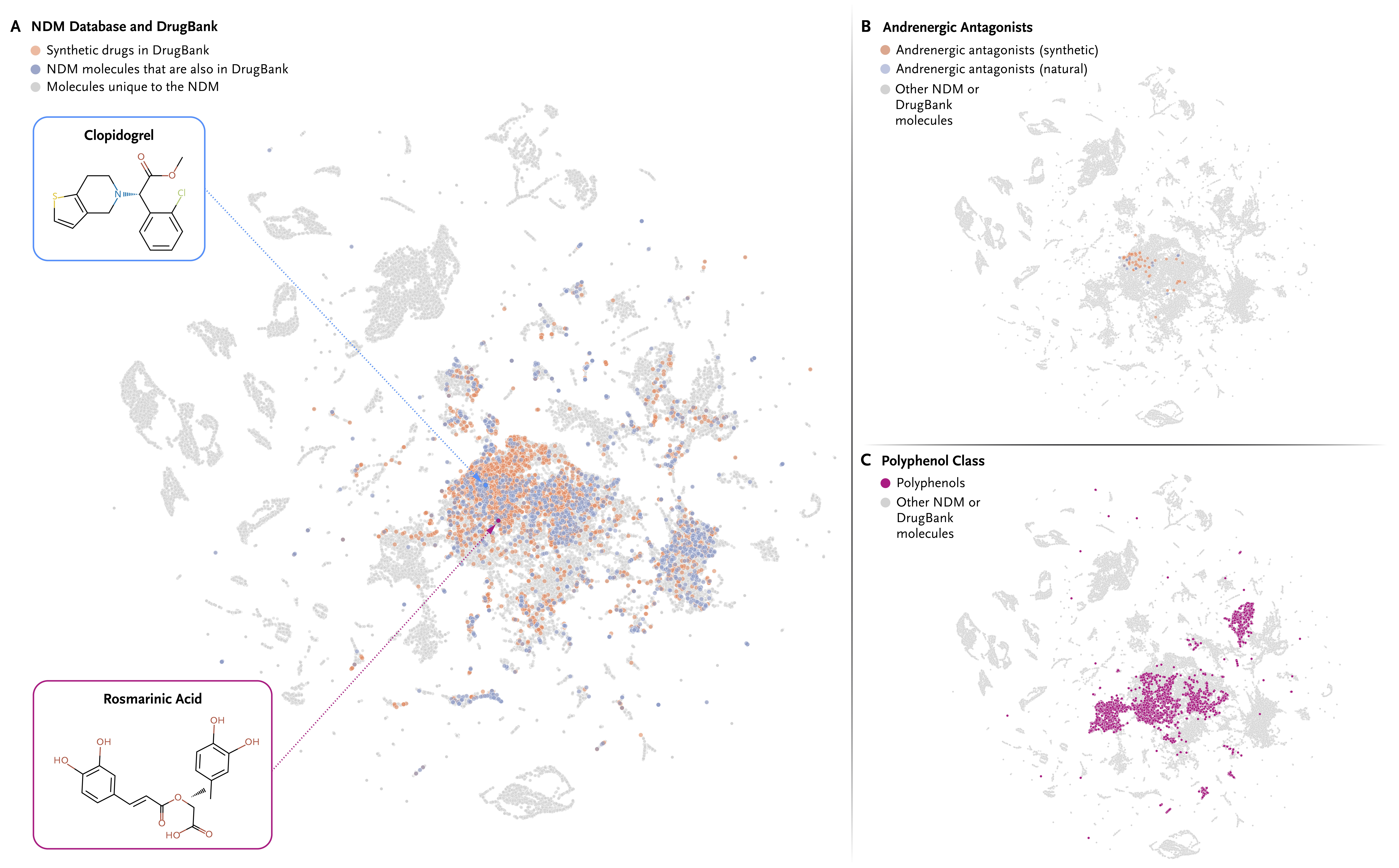
Panel A shows drug compounds clustered in a distinct region of the NDM database. With the use of MoLFormer and UMAP, DrugBank small molecules are visualized within the NDM space. Two antiplatelet agents — rosmarinic acid (a polyphenol) and clopidogrel (a synthetic drug) — coexist in the same neighborhood of the map.
Panel B shows localization of 115 small molecules classified as adrenergic antagonists in DrugBank, either as naturally occurring or synthetic.
Panel C shows that as compared with synthetic drugs, polyphenols have a wider range of structures and features, spreading across a broad region of the chemical space rather than clustering in a narrow neighborhood.
Protein Targets of NDM Mapped onto the Human Interactome.
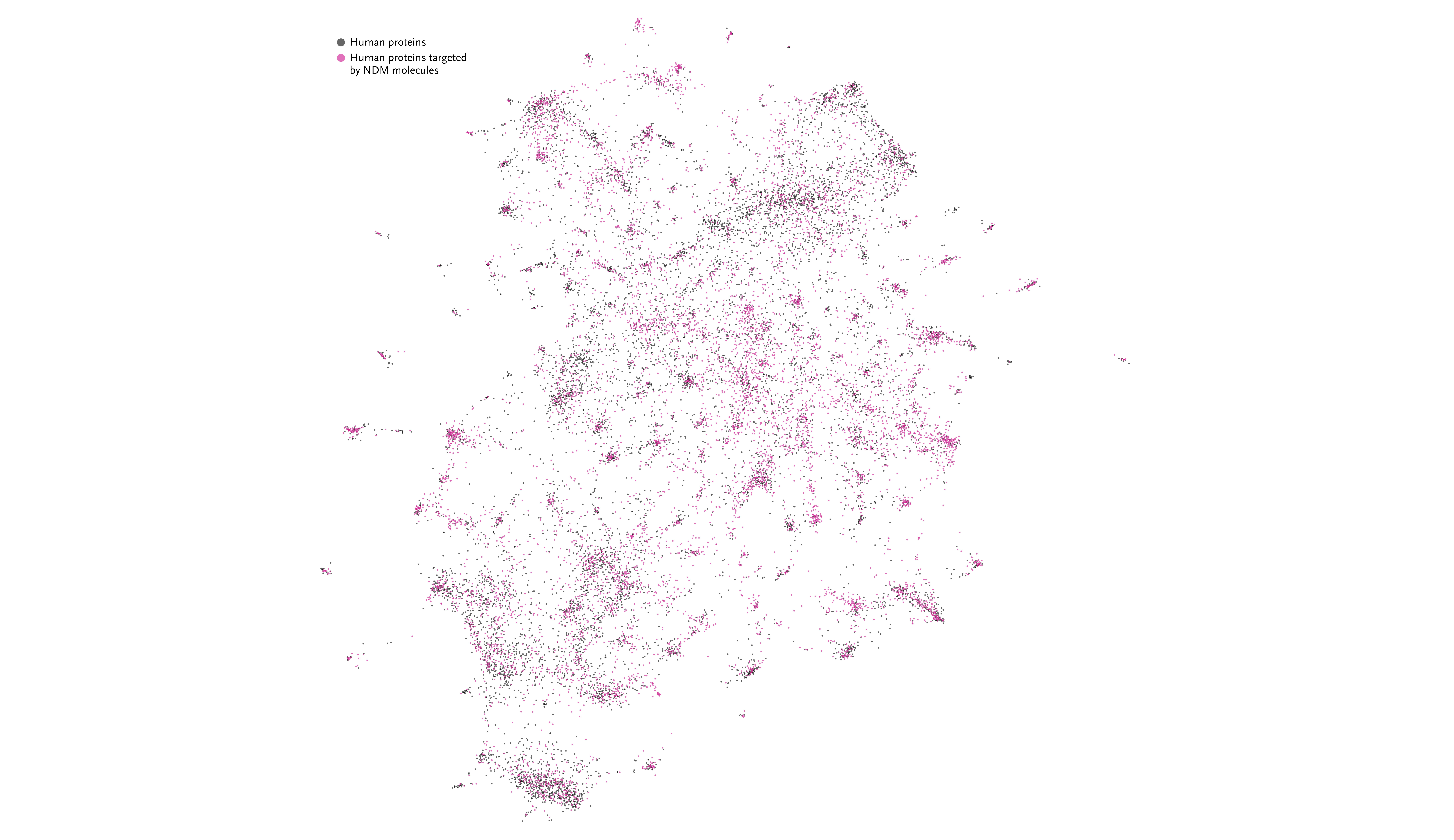
Using node2vec to compress the complex wiring of the human interactome into a 64-dimensional protein-embedding vector, we further apply UMAP to visualize the relative positions of 18,659 human proteins on the basis of their 354,659 physical interactions in a 2-dimensional space. Proteins that frequently co-occur through random walks on the interactome cluster closely together in this space. To facilitate visualization, we have not drawn links between the proteins. Proteins that have experimentally validated binding interactions with small molecules from the NDM library are highlighted in dark pink, revealing that food molecules target nearly half the interactome (8997 proteins).
Resources
Grover A, Leskovec J. node2vec: scalable feature learning for networks. KDD 2016;2016:855-64. [Medline]
Co-Occurrence of Food Molecules and Overlapping Mechanisms of Action.
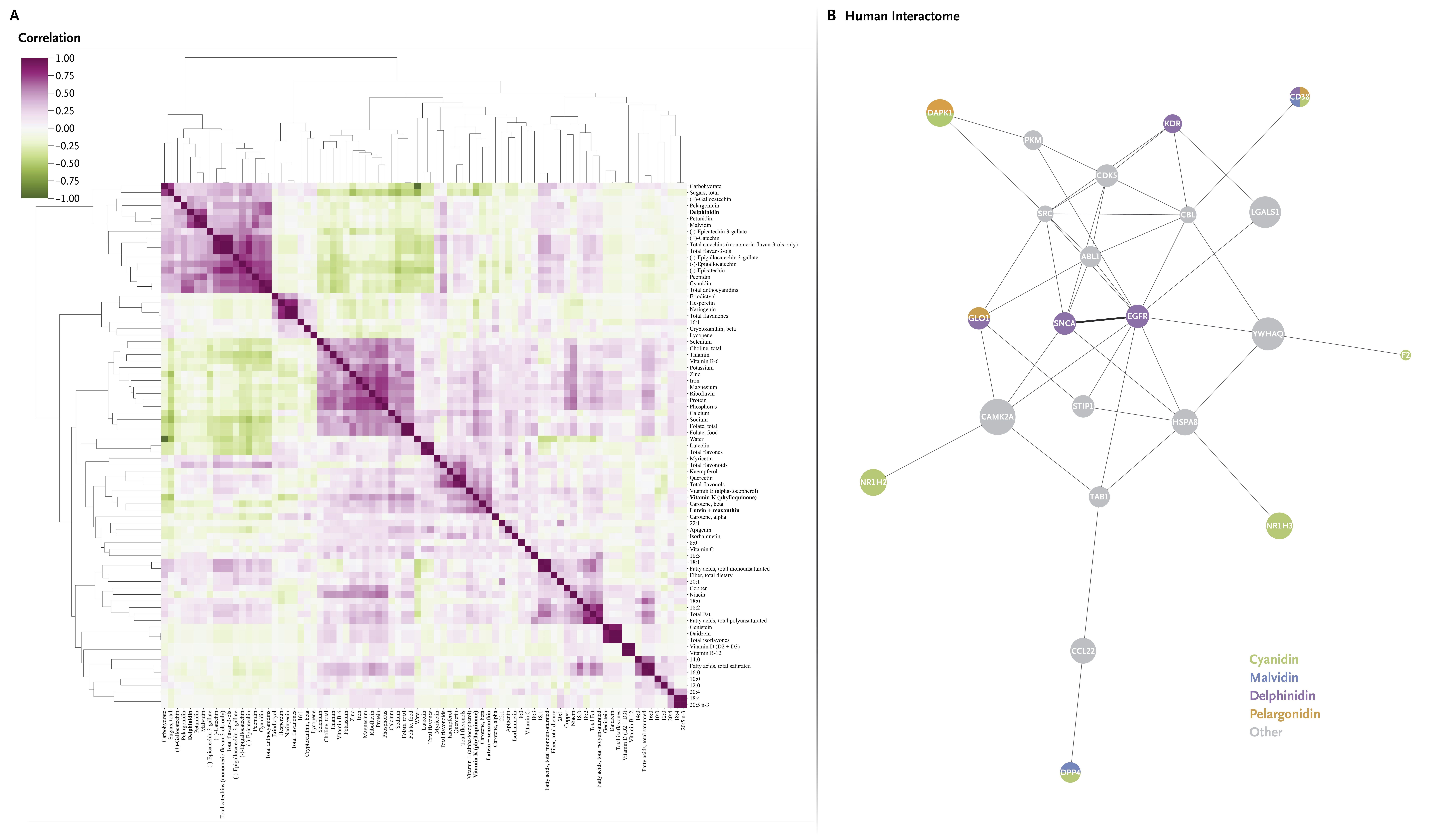
Panel A shows a Spearman correlation matrix of nutrient concentrations for 108 raw fruits and vegetables, profiled with the same resolution by the U.S. Department of Agriculture (USDA). The clusters of highly correlated nutrients indicate compounds likely to have similar production by source plant metabolism. For example, this is the case for vitamin K1 and lutein, which show a high correlation (r=0.6349), despite a difference of at least one order of magnitude in the average concentration. In contrast, delphinidin, an anthocyanidin, belongs to a distinct cluster, indicating no shared biosynthetic pathway. Vitamin K1, lutein, and delphinidin are highlighted in bold.
As shown in Panel B, food molecules produced together tend to target similar regions of the human interactome. Anthocyanidins and anthocyanins, such as delphinidin, cyanidin, malvidin, and pelargonidin, which differ primarily with respect to the presence of sugar groups, share experimentally validated targets within a common neighborhood of the interactome. The 10 protein targets are color-coded on the basis of whether one molecule or multiple molecules bind to them.
Structural Characterization of the Molecules in the NDM Database.
Panel A shows the chemical diversity in the Nutrition Dark Matter (NDM) database. With the use of contextual embedding vectors generated by the MoLFormer chemical language model and reduced to two dimensions with UMAP (Uniform Manifold Approximation and Projection), the space of food molecules in the NDM database can be visualized. Molecules are color-coded on the basis of structural subclasses (ClassyFire chemical taxonomy), each containing at least 500 compounds.
Panel B shows the chemical compounds in garlic. The same map highlights the 6802 molecules documented in garlic, categorized by structural subclasses. Three key chemical compounds — allicin and ajoene (organosulfur compounds) and p-coumaric acid (a polyphenol) — are emphasized for their relevance to human health, yet they are often overlooked in food composition databases.
Resources
Djoumbou Feunang Y, Eisner R, Knox C, et al. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J Cheminform 2016;8:61. [Medline]
McInnes L, Healy J, Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. Version 3. September 18, 2020 (https://arxiv.org/abs/1802.03426). preprint.
Comparison of Molecules in the NDM Database and DrugBank.
Panel A shows drug compounds clustered in a distinct region of the NDM database. With the use of MoLFormer and UMAP, DrugBank small molecules are visualized within the NDM space. Two antiplatelet agents — rosmarinic acid (a polyphenol) and clopidogrel (a synthetic drug) — coexist in the same neighborhood of the map.
Panel B shows localization of 115 small molecules classified as adrenergic antagonists in DrugBank, either as naturally occurring or synthetic.
Panel C shows that as compared with synthetic drugs, polyphenols have a wider range of structures and features, spreading across a broad region of the chemical space rather than clustering in a narrow neighborhood.
Protein Targets of NDM Mapped onto the Human Interactome.
Using node2vec to compress the complex wiring of the human interactome into a 64-dimensional protein-embedding vector, we further apply UMAP to visualize the relative positions of 18,659 human proteins on the basis of their 354,659 physical interactions in a 2-dimensional space. Proteins that frequently co-occur through random walks on the interactome cluster closely together in this space. To facilitate visualization, we have not drawn links between the proteins. Proteins that have experimentally validated binding interactions with small molecules from the NDM library are highlighted in dark pink, revealing that food molecules target nearly half the interactome (8997 proteins).
Resources
Grover A, Leskovec J. node2vec: scalable feature learning for networks. KDD 2016;2016:855-64. [Medline]
Co-Occurrence of Food Molecules and Overlapping Mechanisms of Action.
Panel A shows a Spearman correlation matrix of nutrient concentrations for 108 raw fruits and vegetables, profiled with the same resolution by the U.S. Department of Agriculture (USDA). The clusters of highly correlated nutrients indicate compounds likely to have similar production by source plant metabolism. For example, this is the case for vitamin K1 and lutein, which show a high correlation (r=0.6349), despite a difference of at least one order of magnitude in the average concentration. In contrast, delphinidin, an anthocyanidin, belongs to a distinct cluster, indicating no shared biosynthetic pathway. Vitamin K1, lutein, and delphinidin are highlighted in bold.
As shown in Panel B, food molecules produced together tend to target similar regions of the human interactome. Anthocyanidins and anthocyanins, such as delphinidin, cyanidin, malvidin, and pelargonidin, which differ primarily with respect to the presence of sugar groups, share experimentally validated targets within a common neighborhood of the interactome. The 10 protein targets are color-coded on the basis of whether one molecule or multiple molecules bind to them.